The pandemic of coronavirus disease 2019 (COVID-19) caused by SARS-CoV-2 continues to cause severe health and economic disruptions with far-reaching effects. However, so far, no anti-SARS-CoV-2 drugs or vaccines have been approved, which means that new strategies and rapid solutions are urgently needed. In addition to being busy looking for a vaccine against COVID-19, there is also a very fast ongoing research to find a therapy for SARS-CoV-2. According to different activities, treatment methods can be divided into several categories: (1) preventing the synthesis and replication of viral RNA, (2) blocking the virus from binding to human cell receptors, (3) restoring the innate immunity of the host, (4) Blocking specific receptors or enzymes of the host. Although many exploratory and computational studies have been conducted and all of these categories have been studied, to date, there is no confirmed effective treatment available for COVID-19.
Scientists around the world are intensively searching for inhibitors of SARS-CoV-2. Now, the researchers report on ACS Nano that they have used computer modeling to evaluate four peptides that mimic the viral binding domain of human proteins–a key protein that allows SARS-CoV-2 to enter cells.
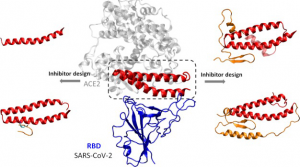
To infect cells, SARS-CoV-2 uses its spike protein to bind to the ACE2 receptor, which is a protein located on the surface of certain human cells. This attachment allows the virus to fuse with the host cell membrane and enter. Many researchers have been trying to find compounds that can block key areas of this spike protein, thereby preventing viruses from infecting cells. Yanxiao Han and Petr Kral wanted to use computer modeling to design compounds that mimic the natural target ACE2 of this spike protein.
Therefore, the researchers examined the x-ray crystal structure of the recently published SARS-CoV-2 receptor binding domain when it binds to ACE2. They identified 15 amino acids from ACE2, which directly interact with viral proteins. Then, the researchers designed four inhibitors that contained most or all of the amino acids and added additional sequences that they thought would stabilize the structure. Through computer simulations, the research team studied how inhibitors bind to proteins in the body and the energy required for binding. One of the compounds has particularly good binding to viral proteins. The research team said that the peptide still needs to be tested in laboratories and patients, but the ability to narrow the range of drug candidates on the computer may help speed up the process.
The inhibitor is mainly formed by two consecutive self-supporting α-helices extracted from the protease domain (PD) of angiotensin-converting enzyme 2 (ACE2), which bind to the SARS-CoV-2 receptor binding domain. Molecular dynamics simulations show that the α-helical peptide retains its secondary structure and provides highly specific and stable binding (blocking) to SARS-CoV-2. To provide multivalent binding to the SARS-CoV-2 receptor, many such peptides can be attached to the surface of nanoparticle carriers. The proposed peptide inhibitor can provide a simple and effective therapy against COVID-19 disease.
Although dozens of research teams around the world are using multiple methods to find new treatments for Covid-19, one of the advantages of this peptide drug is that they are relatively easy to mass produce. They also have a larger surface area than small molecule drugs. Peptides are larger molecules, so they can really hold the corona virus tightly and inhibit entry into the cell, and if small molecules are used, it is difficult to block the entire area where the virus is being used. Antibodies also have large surface areas, so they may also be useful. It just takes longer to make and discover. One disadvantage of peptide drugs is that they are usually not taken orally, so they must be injected intravenously or subcutaneously. They also need to be modified so they can stay in the blood long enough to be effective, and Petr Kral’s lab is working on that.
References:
- Yanxiao Han et al. Computational Design of ACE2-Based Peptide Inhibitors of SARS-CoV-2, ACS Nano (2020). DOI: 1021/acsnano.0c02857