Introduction
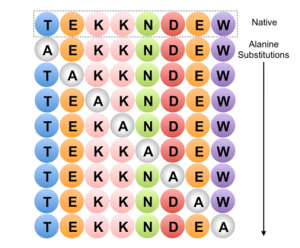
Alanine scanning library is one of the peptide library design tools. Alanine Scanning Library, this tool allows you to systematically substitute alanine for each amino acid position in your peptide sequence (see Fig1). Alanine is the smallest of the naturally occurring amino acids. It can maintain its chirality. In molecular biology, alanine scanning is a site-directed mutagenesis technique used to determine the contribution of specific residues to the stability or function of a given protein. Alanine scanning library can be a powerful tool to explore the effects of single amino acid side chains on protein functions and properties. Alanine can be substituted for each non-alanine residue in turn, one at a time, and subsequently, the corresponding change in epitope activity can be measured. Substituting alanine for key amino acid residues will cause changes in epitope binding activity, so that the contribution of each amino acid to peptide function can be quickly determined.
Figure 1. Example of Alanine Scanning. The native protein (top row) and each possible point mutation to alanine is considered
Fundamental
Alanine scanning mutagenesis is a powerful protein engineering technique used to study protein structure-function relationships, map binding sites, and design more stable proteins or proteins with altered properties. Alanine (Ala) scanning is a widely used mutagenesis method in which residues in the target protein are systematically substituted for alanine at selected positions by site-directed mutagenesis. Alanine is usually the accepted single-residue choice for mutation scanning because it eliminates side chain (R=methyl) interactions, but does not change the main chain conformation, nor does it impose extreme static or steric effects.
Advantages of using alanine
Alanine is used because of its small size, chemical inertness, and methyl functionality, but it mimics the secondary structure preferences of many other amino acids. Therefore, alanine scanning is particularly useful for determining the contribution of specific side chains while preserving the natural and entire protein structure. Sometimes, when the size of the mutated residue needs to be preserved, larger amino acids, such as valine or leucine, are used.
Application field
The alanine scanning library is designed to identify specific amino acid residues responsible for peptide function, stability and conformation.
A good example of alanine scanning is to examine the role of charged residues on the surface of a protein. In a systematic study on the role of conserved charged residues on the surface of the epithelial sodium channel (ENaC), alanine scanning was used to reveal the importance of charged residues in the transport of proteins to the cell surface.
Identify key epitope groups
Alanine scanning is used to simultaneously determine the functional contribution of the 19 side chains buried at the interface between the human growth hormone and the extracellular domain of its receptor. Every amino acid in the side chain is replaced by alanine. Then, a shotgun scanning method that combines the concepts of alanine scanning mutagenesis and binomial mutagenesis with phage display technology is adopted.
Assess the functionally active groups of enzymes
Another key application of alanine scanning is to determine the effect of a single residue on the structure and activity of the prototype cyclic peptide kalata B1. Cyclic peptides display a wide range of pharmaceutically important biological activities, but their natural function is to act as insecticides in plant defense. In the structure of the cyclic peptide kalata B1, all 23 non-cysteine residues are successively substituted by alanine. The data is tested by nuclear magnetic resonance spectroscopy.
References
- Morrison K L, Weiss G A. Combinatorial alanine-scanning[J]. Current opinion in chemical biology, 2001, 5(3): 302-307.
- Weiss G A, Watanabe C K, Zhong A, et al. Rapid mapping of protein functional epitopes by combinatorial alanine scanning[J]. Proceedings of the National Academy of Sciences, 2000, 97(16): 8950-8954.
- Simonsen S M, Sando L, Rosengren K J, et al. Alanine scanning mutagenesis of the prototypic cyclotide reveals a cluster of residues essential for bioactivity[J]. Journal of biological chemistry, 2008, 283(15): 9805-9813.
- Howlader M T H, Kagawa Y, Miyakawa A, et al. Alanine scanning analyses of the three major loops in domain II of Bacillus thuringiensis mosquitocidal toxin Cry4Aa[J]. Applied and environmental microbiology, 2010, 76(3): 860-865.
- Edelheit O, Hanukoglu I, Dascal N, et al. Identification of the roles of conserved charged residues in the extracellular domain of an epithelial sodium channel (ENaC) subunit by alanine mutagenesis[J]. American Journal of Physiology-Renal Physiology, 2011, 300(4): F887-F897.