Non-Ribosomal Peptide Synthetases (NRPSs)
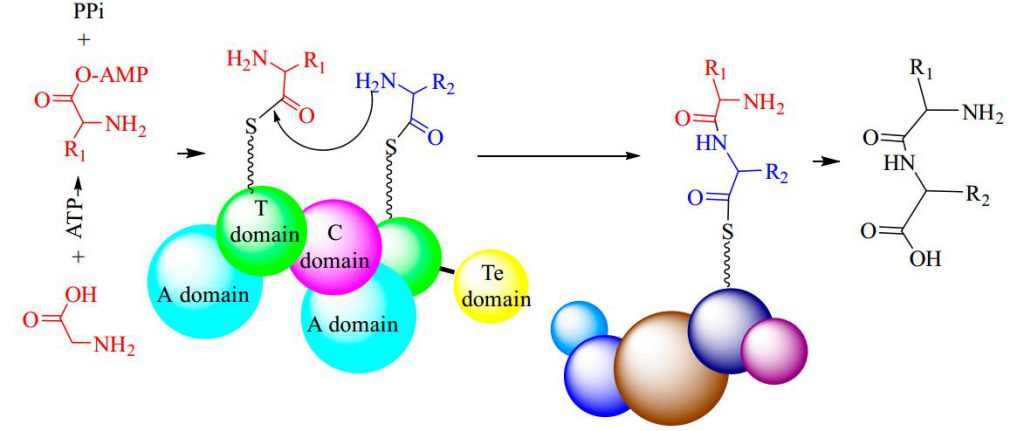
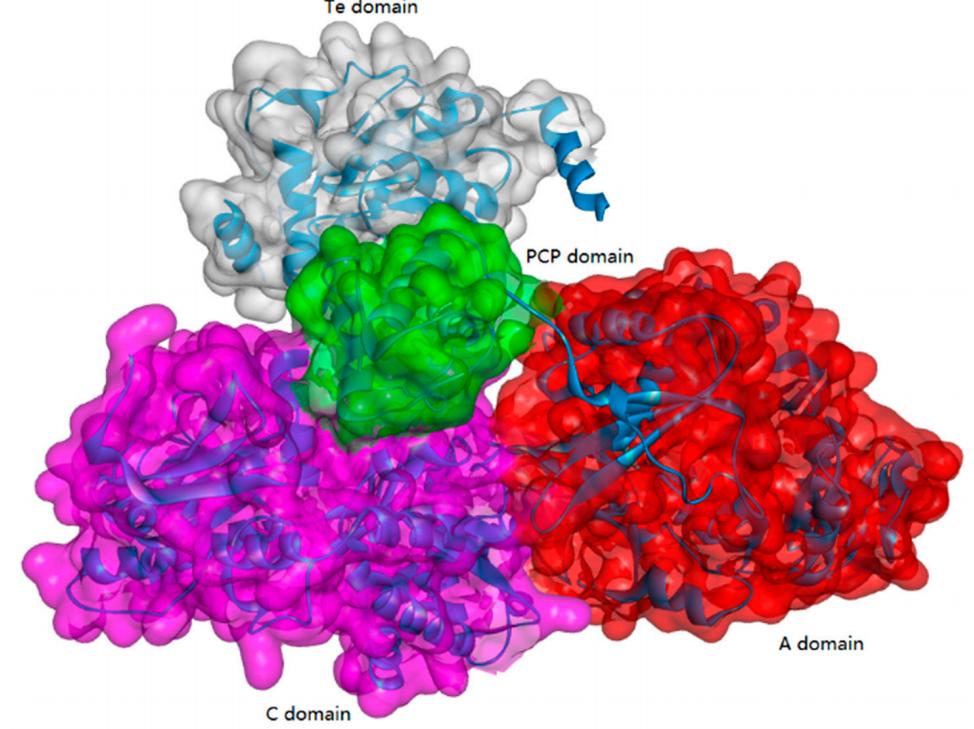
Non-ribosomal peptide synthesis is a universal and critical biochemical process catalyzed by NRPSs in bacteria and fungi, through which a wide array of therapeutically important peptides with highly diverse structures and bioactivities, including penicillin, bleomycin, and cyclosporine, are produced. Biocatalysts capable of NRP synthesis could be divided into two groups (ATP-independent and ATP-dependent enzymes) based on the difference in substrate activation. In the ATP-dependent process, enzymes such as tRNA-dependent ligase can activate the substrate through aminoacyl-adenosine monophosphate. In the ATP-independent process, enzymes such as transacylase use aminoacyl phosphate.
Peptide Synthesis at Creative Peptides
| Services | Description |
| Peptide Synthesis Services | We provide high-quality customized peptide synthesis services, from bulk API peptides, high-throughput library peptides, cosmetic peptides, to array peptides, antigen peptides, and other complex or abnormal peptide sequences. |
| Peptide Modification Services | Creative Peptides offers hundreds of peptide modifications to meet any research need. These modifications can improve the overall stability of peptide, change the structure to better understand biological functions, or enhance the immunogenicity of antibody development and production. |
| Custom Conjugation Service | Creative Peptides provides convenient multiple types of peptide conjugation services to meet the needs of different projects, including activating immune responses, obtaining higher stability and cell penetration. |
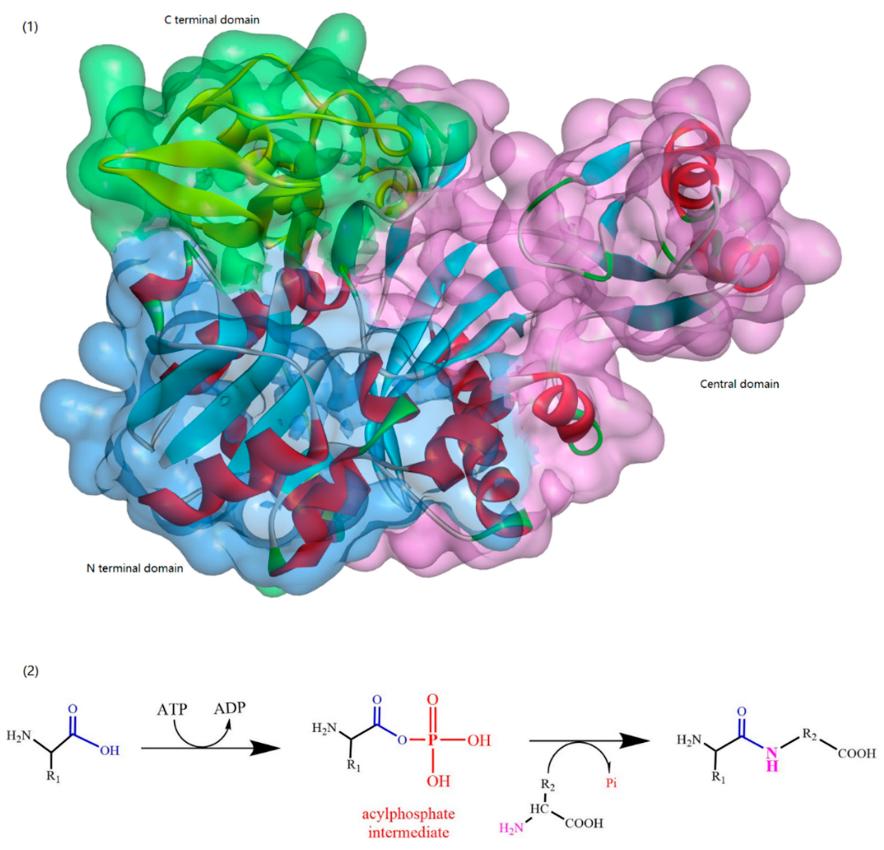
ATP-Grasp Enzymes
The ATP-grasp enzymes, also known as ATP-dependent carboxylate-amine ligases, activate acids like acylphosphate intermediates and are found in various biological systems such as de novo purine biosynthesis. Notable members of this family include biotin carboxylase, Ddl, and glutathione synthetase. ATP-grasp enzymes usually have a unique structure with three conserved domains, with a nonclassical ATP binding fold enclosing an ATP molecule. Most of these enzymes require an Mg2+ ion coordinated by ATP in the active site.
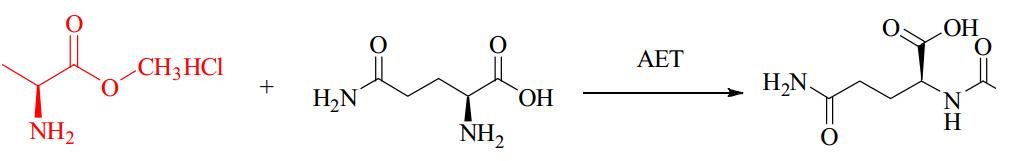
α-Amino Acid Ester Acyltransferase
Kenzo and colleagues reported an efficient enzymatic method for producing oligopeptides from unprotected amino acids with high yield, using Empedobacter brevis ATCC 14234 strain. An enzyme catalyst (named carboxypeptidase Y) was found in the strain to assist in rapid production of certain oligopeptides. However, details about the amino acid sequence, coding gene sequence or 3D crystal structure were not provided. Isao ABE and team first reported about the cloning and expression of α-amino ester acyl transferases (AETs) from the same and another strain, their amino acid sequences were 35% and 36% identical to the α-amino acid ester hydrolase from Acetobacter pasteurianus. AETs displayed dual action-dipeptidyl peptidase and transferase activities, and was quite specific for both acyl donors and nucleophiles. There have been no studies yet on the 3D structure of AET or its reaction mechanisms.
Enzymes Used for β-Lactam Biosynthesis (β-Lactam Acylases)
β-Lactam antibiotics, which contain a β-lactam ring in their chemical structure and include types such as penicillin, cephalosporins, and thiamycins, are widely used as anti-infective agents and have significant importance in the pharmaceutical industry. The environmentally-friendly and cost-effective synthesis of β-lactam antibiotics via enzymatic means has been increasingly applied in modern pharmaceutical enterprises. β-lactam acylases, which historically process β-lactam antibiotics, can also be used for the creation of semi-synthetic β-lactam antibiotics. Different types of β-lactam acylases such as penicillin acylase (PA), glutaryl acylase (GA), and β-amino acid ester hydrolase (AEH) have been studied in the biosynthesis of β-lactam antibiotics. PAs are generally produced by a variety of microorganisms and are categorized into two types based on their substrate specificity. They are widely used in the industrial production of the active pharmaceutical intermediate 6-aminopenicillanic acid (6-APA) and for the synthesis of semi-synthetic antibiotics, making them potentially useful in the development of novel drugs. Furthermore, PAs can be used in peptide synthesis, resolution of racemic mixtures, and in the production of both achiral and chiral compounds for pharmaceutical intermediates.
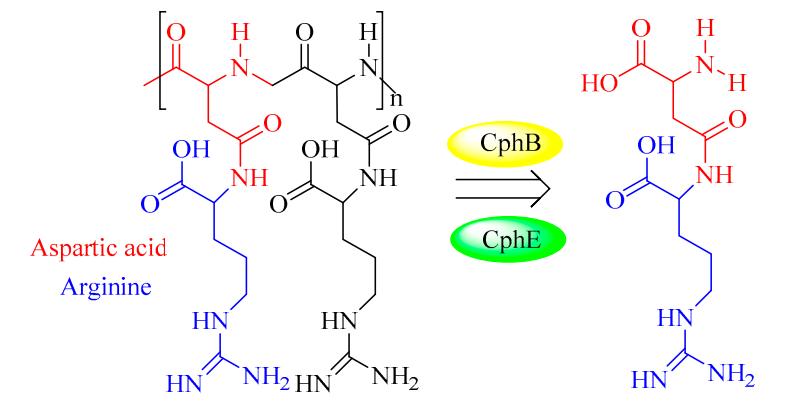
Cyanophycinases (CGPases)
Cyanophycin granule polypeptide (CGP, or multi-L-arginyl-poly) is an intracellular storage polymer found in most cyanobacteria. Equimolar concentrations of arginine and aspartic acid are observed in the aspartic acid backbone, where the arginine moieties are linked to the β-carboxyl group of each aspartic acid through its α-amino group. In most genera of cyanobacteria, the cyanophycin synthetase gene (cphA) has been identified and verified for the synthesis of CGP. In contrast, the intracellular and extracellular degradation of CGP is catalyzed by cyanophycinases (CphB and CphE), which releases dipeptides (β-Asp-Arg). In this respect, β-Asp-Arg can be efficiently synthesized via the simultaneous production of CGP and CGPase, which could be further applied in various fields requiring arginine (Arg) content in feed or food. However, the production and efficient isolation of CGP from various organisms have been successfully established in several recombinant strains, including Escherichia coli, Nicotiana tabacum, Pseudomonas putida, and Pseudomonas alcaligenes DIP1. Therefore, it is very feasible to produce dipeptides (e.g., β-Asp-Arg) via the metabolic engineering of suitable hosts and chemo-enzymatic strategies. For example, successful co-expression of CGP and CGPase in the Nicotiana tabacum plant was recently achieved A further study showed that it is possible to realize the goal of sufficient storage and efficient transport of arginine and β-Asp-Arg dipeptides in this synthetic model.
Reference:
- Wang, T., et al. Strategy for the biosynthesis of short oligopeptides: Green and sustainable chemistry. Biomolecules. 2019, 9(11): 733.