Cyclic peptides can be produced chemically by using solid-phase peptide synthesis, enzymatic chemistry, or expressed in microorganisms using DNA recombinant techniques.
Cyclic peptides ranging from approximately 15-40 amino acids long can be readily synthesized using solid-phase peptide synthesis (SPPS). It involves two steps process: production of linear precursor and cyclization.
Linear precursor α-thioester peptides can be produced by both Boc-based and Fmoc-based chemistry. The most commonly used approach for the production of -thioester linear cyclotide precursors is using the 3-mercaptopropionamide linker (Boc-based), sulfonamide linker and 3,4-Diaminobenzoic (Dbz) linker (Fmoc-based).
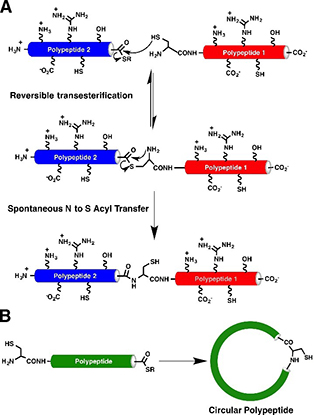
The most commonly used method for the backbone cyclization of the linear cyclotide precursor employs an intramolecular NCL, in which the peptide sequence contains an N-terminal cysteine and an α-thioester group at the C-terminus (Fig. 1A).
Cyclization can be formed by direct chemical coupling of the Nα-amino and C-terminal carboxylic groups using uronium salts as coupling reagent.
Also enzymes such as proteases and trans-peptidases can be used for the production of backbone cyclized polypeptides.
Synthesis of backbone-cyclized polypeptides can be also produced using standard heterologous expression systems such as E. coli.
An intramolecular version of intein-mediated ligation (also called Expressed Protein Ligation (EPL)) can been used for the generation of many different types of backbone-cyclized polypeptides and proteins. It utilizes molecular cloning of DNA encoding linear precursors into heterologous expression systems. Once the linear precursor is expressed, it is cyclized via intermolecular NCL to form cyclic peptides (Fig. 1B).
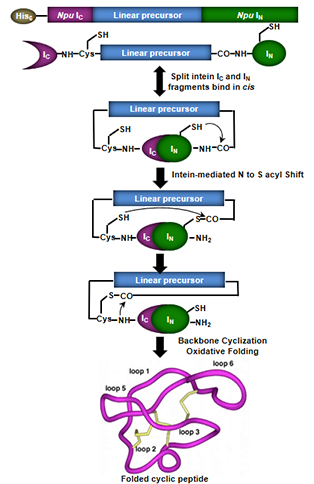
Protein trans-splicing is another way of producing cyclic peptides. It is a post-translational modification similar to protein splicing with the difference that the intein self-processing domain is split into two fragments (N-intein and C-intein). The split-intein fragments are not active individually, however, they can bind to each other with high specificity under appropriate conditions to form an active protein splicing or intein domain in trans. By rearranging the order of the intein fragments, i.e. by fusing the N- and C-intein fragments to the C- and N-terminus of the polypeptide to cyclize, the trans-splicing reaction results in the formation of a backbone-cyclized polypeptide (Fig. 2).
Backbone-cyclized peptides are peptides with a covalent bond between their N- and C-termini, forming a cyclic structure. They can be synthesized chemically using solid-phase peptide synthesis (SPPS) or biologically via recombinant DNA techniques.
SPPS involves synthesizing a linear peptide, followed by cyclization through chemical or enzymatic methods, such as using uronium salts or native chemical ligation (NCL).
Cysteine is crucial for cyclization, especially in chemical methods, where its N-terminal group forms a bond with a C-terminal thioester group, enabling peptide cyclization.
Enzymatic methods, such as using proteases, offer high specificity and control over the cyclization reaction, leading to higher purity and fewer side reactions.
Yes, recombinant DNA techniques express linear peptide precursors in systems like E. coli, which then undergo enzymatic cyclization, such as expressed protein ligation (EPL).
EPL involves expressing linear peptides in a host system, which are then cyclized via intermolecular native chemical ligation (NCL), enabling efficient peptide synthesis.
Protein trans-splicing uses split inteins to catalyze the cyclization of peptides by binding the N- and C-intein fragments together, forming a cyclic structure.
Disulfide-rich cyclic peptides offer enhanced stability, making them ideal for drug discovery and biomolecular applications due to their resistance to proteolysis.

References