Peptide library is a collection of a large number of small peptides with specific length and different sequences, which includes the arrangement and combination of various (or most) amino acid sequences in short peptides with this length. Although some peptides have different sequences from the natural epitopes of antigens, they bind to antibodies or ligands in the same way, so this kind of peptide with key amino acid residues is called mimotope. The concept of mimic epitope has played an important role in the development of peptide library. Peptide library can provide powerful tools for drug design, protein-protein interaction and other biochemical and drug research and application.
Creative Peptides is devoted to revolutionizing the medical and healthcare sector, offering an exclusive collection of peptide libraries. Our company prioritizes peptide-based drug discovery, paving the way for breakthroughs in understanding the biological and therapeutic roles of peptides.
Combinatorial peptide libraries are large collections of peptides that are systematically synthesized in order to generate a variety of sequences. This approach uses principles of combinatorial chemistry, which involves creating a large number of compounds by reacting a set of building blocks in all possible combinations. The result is a library of peptides, each with a unique sequence, that can be screened for various properties, such as binding capacity to a target protein. These libraries can be very diverse, potentially containing millions of distinct peptide sequences.
Combinatorial peptide libraries can be categorized into two groups-chemical peptide libraries, which are produced via organic synthesis, and biological libraries.
Various screening or selection methods are available based on the type of peptide library platform. Common procedures include using a fluorescently labeled target or target-coated magnetic beads, combined with flow cytometry-based systems like FACS, or magnetic separation techniques like MACS. The former is mostly used for cell-based peptide libraries, although it has also been used for screening chemical library systems such as one-bead-one-compound platform. Identification of hits is reliant on the type of library; iterative deconvolution or positional scanning methods are used for chemical libraries, while sequencing is used for DNA-encoded platforms. Recently, next-generation sequencing (NGS) methods have advanced library screening by detecting low abundant clones and quantifying changes in clone copy numbers without the need for multiple rounds of selection.
In chemical approach, the library synthesis is either completed on a solid support (insoluble porous polymeric resin) and then members are cleaved to be screened as free compounds, or the library is synthesized and screened on a solid matrix. By randomly synthetic peptides on the resin, a peptide library of thousands of peptides can be synthesized at one time. For example, a five-peptide library will have five pentapeptides to the fifth power, and a nine-peptide library will have nine nonapeptides to the ninth power.
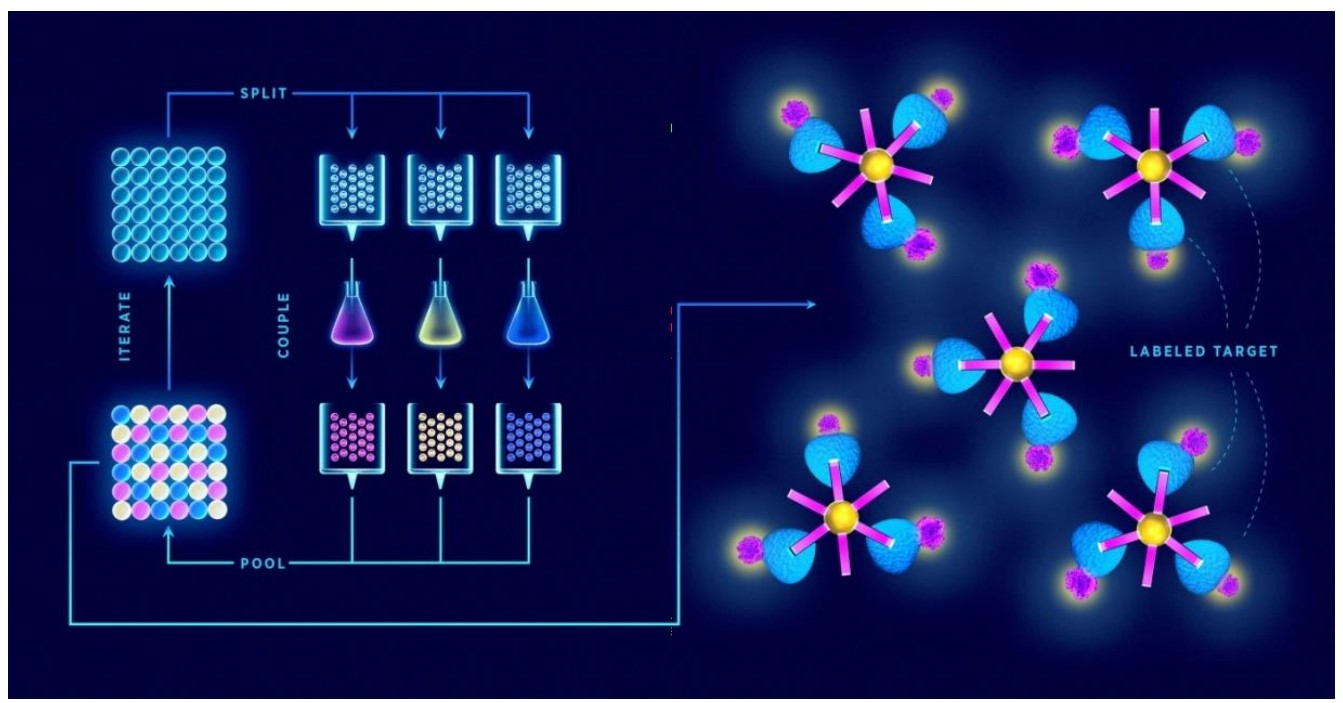 Fig. 1 Synthesis of the name "one-bead-one-compound" combinatorial peptide libraries by the "split-and-mix" method. (Bozovičar, K., 2019)
Fig. 1 Synthesis of the name "one-bead-one-compound" combinatorial peptide libraries by the "split-and-mix" method. (Bozovičar, K., 2019)
A key feature of biological libraries is the linkage of the genotype (i.e., genetic information) with its corresponding phenotype (the encoded peptide/protein). In directed evolution, (random) mutagenesis (gene diversification) and screening for functional gene products are iteratively alternated, and the phenotype: genotype link is crucial for peptide identification via sequence determination of the encoding oligonucleotide. Based on the display type approach, biological libraries can be categorized as either cellular or acellular.
Creative Peptides provides a variety of peptide libraries for the separation of minimal length active peptide sequences, identification of critical amino acid residues, design of sequence-optimized analogs, etc. For protease and kinase studies, we also supply peptide microarrays. Meanwhile, we providing relevant analytical services to meet customer needs.
The design of overlapping peptide library is mainly determined by two parameters: peptide chain length and offset number. The design is gradually intercepted from the N-terminal to the C-terminal of the target protein, and one or several amino acids are transferred at a time, and the sequences partially overlap, and finally an overlapping peptide library is formed. It is mainly used for whole protein scanning and provides an ideal tool for screening linear or continuous epitopes. According to the target protein or long peptide, the key short peptide sequence can be used to identify the protein activity, immune specificity and antibody binding.
Used to determine the minimum range of epitopes. The method of building truncated peptide matrix is generally obtained by systematically removing amino acids on both sides of the original peptide sequence. The head peptide matrix is used to identify the minimum amino acid sequence related to activity.
The library is used to identify specific amino acid sites closely related to the function, stability and conformation of peptides. Alanine (Ala) is the smallest chiral amino acid. By systematically and sequentially replacing each amino acid in the sequence, the influence of a specific amino acid on the biological activity of the whole protein can be distinguished.
A particular site in the peptide sequence is replaced by other different amino acids in the system to study the role of a particular position in replacing the amino acid. By determining the corresponding increase in activity, the optimal amino acid at these sites can be determined.
It is the peptide library with the highest degree of mutation, which is formed by rearranging and combining fragments of the original sequence. Random library can display all possible peptide substitution structures, which is an ideal means of sequence optimization and can be used as targeted molecular probes for proteins, antibodies, genes and so on. Random peptide library can be used as negative control
The unique sequence generated by random peptide library has the potential to enhance peptide activity. This kind of peptide library is obtained by random and spontaneous substitution of 20 natural amino acids at selected sites by shotgun approach.
Peptide array (or peptide chips) is a collection of peptides displayed on a solid surface. Peptide chips are widely used in drug research and development, clinical testing, and basic research in life sciences. In the field of basic research, peptide chips are usually used for the characterization of enzymes (such as kinases, phosphatases, proteases, etc.), the identification of epitope determinants of antibodies, and the identification of key groups that bind to proteins. In the field of clinical application, peptide chips are often used in the discovery of blood markers, the changes of humoral immune response of patients in the process of disease, curative effect monitoring, patient grading, diagnostic tools development and vaccine development.
Shah et al. analyzed the specificity of 52 commercial antibodies in distinguishing different methylation forms (Me1, Me2, Me3) of histone H3K4 by using histone peptide array chip and internal calibration chip. As we all know, post-translational modification of histone (PTM) is a genomic regulatory factor often studied in chromatin immunoprecipitation (ChIp) experiments. Its position and relative abundance are inferred by ChIp, but it will be influenced by the antibody specificity of DNA and histone complex. The Qualcomm capacity, high efficiency and specificity of microarray peptide chip can meet the screening requirements of multiple antibodies. In the experiment, the researchers combined peptide array and ICeChIP analysis to critically evaluate antibodies, and screened out high affinity antibodies with different methylation forms of histone H3K4.
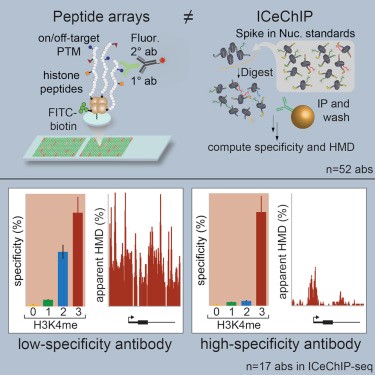 Fig. 2 Peptide chips are used to screen high affinity antibodies that bind to different methylated forms of histone H3K4. (Shah, R. N., 2018)
Fig. 2 Peptide chips are used to screen high affinity antibodies that bind to different methylated forms of histone H3K4. (Shah, R. N., 2018)
Peptide library is a powerful screening tool in biological and chemical research. It is used to screen a very small number of peptides with key biological activities from a large number of peptides. Peptide libraries are widely used in protein omics and its related fields, such as drug research and development, protein-protein interaction, epitope screening, GPCR ligand screening, protein function analysis, enzyme substrate or inhibitor screening, messenger molecule development and peptide/protein signal response.
Using T cells to target and eliminate viruses and cancer cells has become a very important research field. Immunotherapy becomes more effective by engineering receptors to recognize specific TAAs or virus epitopes. Chimeric antigen receptor T cell is an example of this kind of cell, and peptide library is very valuable for identifying the antigen sequence of T cell receptor.
Peptide library is usually used to identify immunogenic epitopes and evaluate the effectiveness of vaccines. Since the library can span the whole protein sequence, it will not miss any important epitopes. However, for some highly infectious diseases, peptide library is an ideal tool for in vitro immune monitoring.
Targeted therapy is the key to effectively eliminate cancer cells, and peptide libraries are usually used to quickly identify important biomarkers on cancer cells. Peptides corresponding to tumor-associated antigen (TAA) were displayed in the form of library, and bioactive peptides were screened out. Once the peptides with specific activity are identified, highly specific antibodies can be designed for cancer immunotherapy or cancer vaccine.
Identifying antigens that stimulate the binding of antibodies to T cells is an important first step in the development of immunotherapy. Potential antigens are sequenced and separated to form overlapping peptides. When the binding of positive T cells or antibodies is confirmed, these peptide sequences can be used to design targeted therapy.
Peptide library has been widely used in protein omics research, such as screening proteolytic peptides or immune monitoring. These libraries are usually combined with mass spectrometry (MS) to screen important biomarkers or identify post-translational modifications.
Peptide libraries are ideal tools for many biological experiments. These libraries can be used to identify peptides with antibacterial activity, direct stem cell differentiation and even monitor pharmacological activity.
Extensive range: We offer an extensive range of Peptide libraries, suited to various research and development needs.
High-quality products: We provide high-quality, well-characterized peptide libraries and arrays which undergo strict quality control.
Peptide library design: We offer customization options, allowing you to tailor your peptide library to fit your specific research requirements.
Expertise: Our team comprises seasoned scientists with vast experience and technical expertise in the field of peptide library synthesis.
Customer support: We provide excellent customer support, assisting you throughout the order process and beyond.
Fast turnaround time: Our efficient processes and dedicated team ensure quick order fulfillment without compromising on quality.
Peptide arrays are solid surfaces to which a library of peptides is attached in a grid-like fashion. These arrays are employed in high throughput screenings to determine interactions between compounds and proteins or to profile immune responses.
Our peptide libraries and arrays are widely applied in the field of high-throughput epitope mapping, protein-protein interaction, vaccine development, drug discovery, and many other fields of quality control.
We offer multiple formats including plate-based libraries, bead-based libraries, as well as custom arrays based on client's research needs.
Our peptide libraries and arrays are synthesized with the most advanced technology ensuring high purity and quality. They are then stringently validated to ensure the accuracy and reproducibility of your assay results.
Typically, it takes around 3-4 weeks to synthesize a peptide library or array. However, the actual time can vary depending on the complexity of the sequences and the current workload.
Our peptides and peptide arrays are carefully packaged and shipped on dry ice. Upon delivery, they should be stored at -20°C for short periods or at -80°C for long term storage to ensure their stability.
You can directly contact our sales representatives or submit a request form on our official website to order. We will need information about your desired sequences and the format you prefer, along with any other specificities you require.

References